Biodegradation
Biodegradation refers to the transformation of a chemical by enzymatic reactions in micro-organisms. In soil and sediment, biodegradation is often the most important factor in the removal of the chemical from the environment. Depending on the ambient conditions, different modes and rates of biodegradation may predominate and may make a chemical readily biodegradable at one site, but not at another, largely due to different degradation capacities of the surrounding environment. In general, microbial transformation is the main mechanism by which an organic chemical can be completely mineralised. In Chapter 2 a review of various OECD biodegradation test systems are summarised, and their applicability to ionisable organic compounds is discussed. This Section briefly reviews some of the more common estimation methods used for biodegradation.
Like other estimation methods, approaches to estimate biodegradation can be categorised according to the development method. The most common approaches are chemical class-specific regression models, expert systems (including probabilistic models) and hybrid approaches. A recent review by (Pavan and Worth, 2008) provides a more thorough review of the various estimation methods. Complementing the biodegradation review is the summary of biodegradation removal of pharmaceuticals in waste water treatment systems by (Onesios et al, 2009), which provides an indication of some of the challenges associated with estimating removal by biodegradation of the APIs that are highlighted in this report.
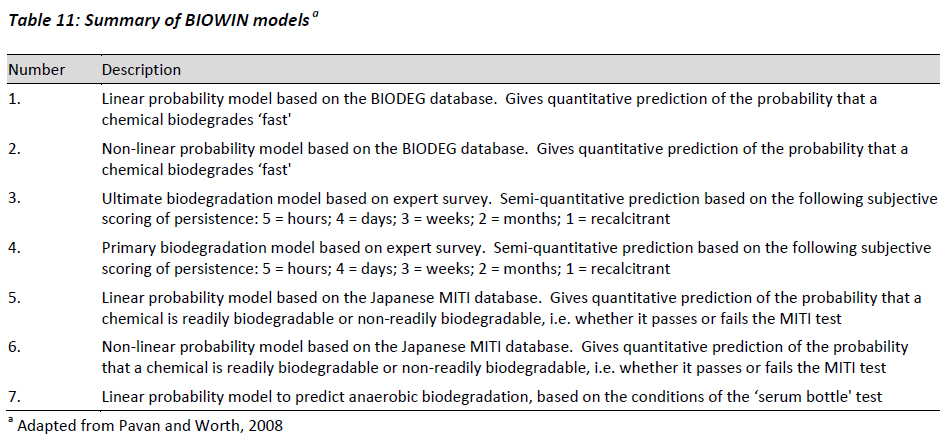
BIOWIN
The biodegradation probability program (BIOWIN) represents one of the most widely used estimation method for the biodegradation of general chemicals (Pavan and Worth, 2008). The program was developed by the Syracuse Research Corporation on behalf of the US EPA, and is freely available for download at http://www.epa.gov/oppt/exposure/docs/episuitedl.htm. There are currently seven different modules included in the BIOWIN program that include both linear and non-linear regression models based on 36 pre-selected fragments and molecular weight. Table 11 summarises each of the different models.
The BIOWIN model is used as a tool within the EU TGD to assess persistence when no data are available for a particular substance, or the available data are difficult to interpret (EC, 2003). The EU TGD recommends the use of the BIOWIN2 model output < 0.5 or BIOWIN6 model output < 0.5 and BIOWIN3 output . months (< 2.2) should reliably indicate a substance that will not readily biodegrade, and therefore has the potential to be persistent in the aquatic environment EC, 2003). In general the recommendations present in the EU TGD reflect the performance of the BIOWIN models, in that they typically perform well when predicting non-readily biodegradable substances better than predicting readily biodegradable substances.
A shortcoming of the BIOWIN models is that they utilise a fragment approach, and therefore poorly account for the influence of neighbouring substituents and substituent position (ref BIOWIN programme). Two examples for which known problems exist are chlorinated phenols and napthoic acids. Other problems with the approach are related to the summation of fragments, no matter what their type or number, which can work well when the molecule in question is small but leads to errors when the frequency of the fragment in the molecule is relatively large. Additionally, not all fragments of significance for biodegradation are included, and it is acknowledged that BIOWIN lacks fragment coefficients for phosphonate; imidazole ring; pyrimidine ring; ethoxylate ether; cycloaliphatics; quaternary nitrogen (except BIOWIN7 which does have this fragment); and others. If a chemical contains a unique or unusual substructure not included in the model's fragment library, these features will not be considered in the prediction process. About 20% of the APIs included in Appendix F include either an imidazole or pyrimidine ring, and in some instances both. Nevertheless, based on the output from the BIOWIN programme, the majority of substances listed in Appendix F would be classified as non-readily biodegradable, with only 5 chemicals being identified as readily degradable (phenyephrine; piperazine; pregabalin; zanamiver; mupirocin). The criteria for predicting readily degradable is if the BIOWIN3 result is eweeks' or faster and the BIOWIN5 probability is . 0.5. This observation is in general agreement with empirical data that attempt to measure the removal efficiency of APIs in waste water treatment systems, where it is widely acknowledged that the dominant removal mechanism is via sorption to suspended organic matter that is subsequently removed by coagulation (Oulton et al., 2010).
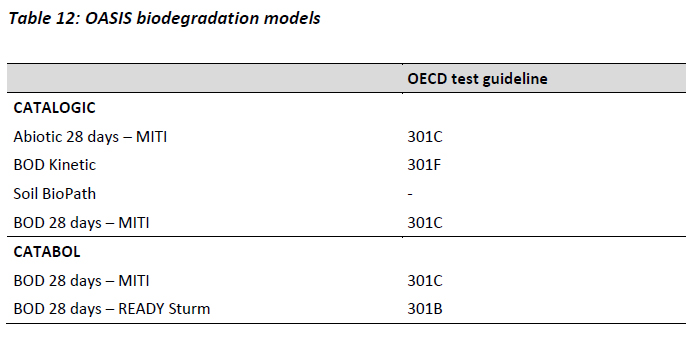
OASIS
The most recent version v5.11.1 offers the user two main fields of applicability namely biodegradation and bioaccumulation, both of high relevance regarding the hazard assessment of chemical substances. However, further detail regarding the relevance of the bioaccumulation model for ionisable organic compounds requires further investigation, as this represents an area of emerging research.
The biodegradation models are separated into two sub-groups: CATALOGIC models which apply metabolic pathways and kinetic information on transformation and the so called pioneer models of CATABOL which are not accounting for such techniques. (Table 12).
A mechanistic approach is used in CATALOG for modelling metabolism. This includes a library of known enzymatic reactions. Each enzymatic reaction is defined as a specific biotransformation. Within each of these specific processes a chemical functional group or a sub-structural fragment of a molecule is modified by the specific enzyme. As a result, the respective metabolic derivative is formed. Subsequently, metabolic pathways are predicted by applying a recursive method that identifies the individual biotransformation paths which are possible for the target chemical. Then every metabolite again is treated as a new target chemical to create the metabolite tree which consists of different metabolic pathways of a different probability each. This recursive application of the biotransformation rules allows a reproducible generation of a metabolic tree.
While the metabolic tree is easily reproduced, there are limited data sets that establish its relevance to what's actually observed in the environment. Recent work by Ericson (2012) using the University of Minnesota Biocatalysis Database and Profiling System (Gao et al, 2010) noted that many of the predictions shown for pharmaceuticals do not match up with what's seen in the OECD 308 water-sediment biotransformation test; or requiring extensive filtering through the predictions to find the observed transformation product. It is thought that the rules in the prediction system may have not used pharmaceuticals in their training sets and/or have not accounted for some of the unique chemistries used in pharmaceuticals to provide desired pharmacokinetics, distribution and/or elimination.
Consistent with the output from the BIOWIN models, the majority of substances listed in Appendix F would be classified as non-readily degradable based on output from the BOD 28 days MITI (301C) model. Seven substances, however, have a projected biodegradation of > 60% after 28 days, those being albuterol, carvedilol, fluorouracil, naratriptam, salmeterol, phenylephrine, and pregabaline, where the latter two were also identified as readily degradable by BIOWIN.